- Home
- Resource
- Explore & Learn
- Enzyme-Driven Nucleic Acid Amplification for Molecular Diagnostics
- Home
- IVD
- By Technology Types
- By Diseases Types
- By Product Types
- Research
- Resource
- Distributors
- Company
Molecular diagnosis has revolutionized the field of healthcare by enabling rapid, sensitive, and specific detection of infectious agents, genetic mutations, and various diseases. At the heart of molecular diagnosis lies Nucleic Acid Amplification Technologies (NAATs), which are essential for amplifying minute quantities of DNA or RNA to detectable levels. These technologies rely heavily on the activity of specialized enzymes that catalyze the replication and amplification processes. This comprehensive guide delves into the critical role of enzymes in NAATs, exploring their mechanisms, applications, and advancements.
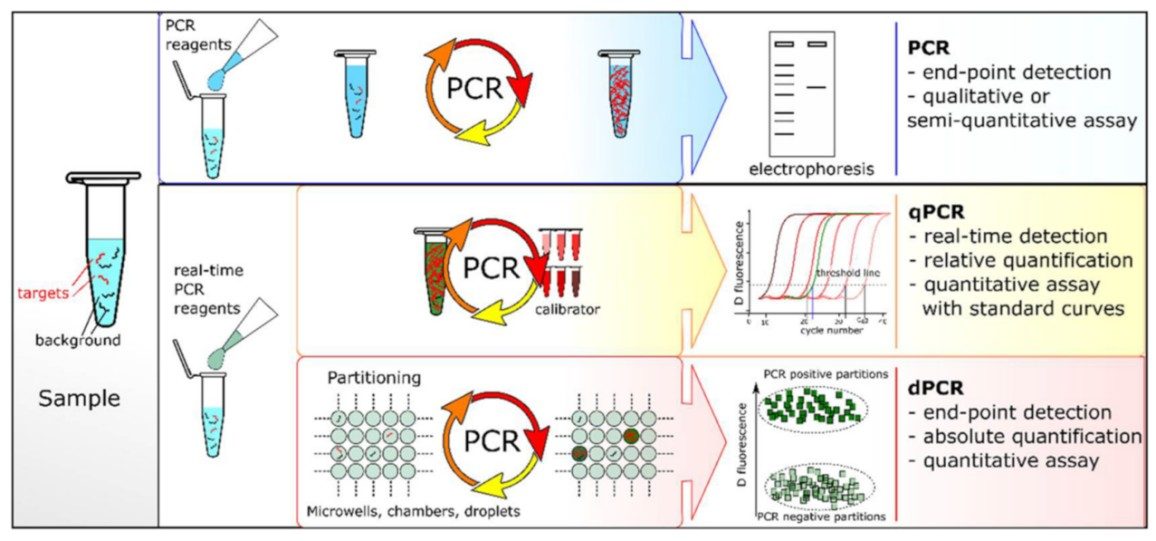 Fig.1 PCR comparisons: end-point PCR, qPCR and dPCR. (Wang M., et al., 2023)
Fig.1 PCR comparisons: end-point PCR, qPCR and dPCR. (Wang M., et al., 2023)
Loop-Mediated Isothermal Amplification (LAMP)
LAMP, invented by Notomi et al. in 2000, is a prominent isothermal amplification technique that operates at a constant temperature (60-65°C). LAMP employs Bst DNA polymerase, which possesses strong strand displacement activity, eliminating the need for thermal cycling. The reaction utilizes four to six primers designed to recognize six distinct regions of the target DNA, ensuring high specificity. LAMP's rapid amplification and high tolerance to inhibitors make it ideal for point-of-care testing in resource-limited areas.
Recombinase Polymerase Amplification (RPA)
RPA, developed by Piepenburg et al. in 2006, is another innovative isothermal amplification method that operates at 37-42°C. RPA relies on three key enzymes: recombinase (T4 UvsX), single-stranded DNA binding protein (SSB), and Bsu DNA polymerase. The recombinase forms a protein-DNA complex with primers, facilitating their hybridization to the target DNA. SSB stabilizes the displaced strand, while Bsu polymerase extends the primer, synthesizing new DNA strands. RPA's rapid detection capabilities and compatibility with fluorescent probes make it suitable for real-time diagnostics.
Rolling Circle Amplification (RCA)
RCA, developed by Salas and colleagues in 1989, is based on the principle of rolling circle replication observed in circular plasmids and virus genomes. RCA utilizes phi29 DNA polymerase, known for its high processivity and strand displacement activity. The enzyme synthesizes long, single-stranded DNA concatemers, enabling high-sensitivity detection. RCA's versatility extends to the detection of both DNA and RNA targets, making it valuable in various diagnostic applications.

NAATs have transformed the landscape of infectious disease diagnosis by enabling rapid and accurate detection of pathogens. For example, RT-PCR remains the gold standard for diagnosing COVID-19, while isothermal amplification methods like LAMP and RPA offer on-site testing capabilities. The integration of CRISPR-Cas systems further enhances the sensitivity and specificity of these assays, facilitating early detection and timely intervention.

Early diagnosis of cancer significantly improves patient outcomes. NAATs play a crucial role in detecting circulating tumor cells (CTCs) and tumor-derived nucleic acids in bodily fluids. RCA, in combination with electrochemical or fluorescent sensors, enables highly sensitive detection of CTCs. CRISPR-based assays, such as CRISPR-Cas12a, offer rapid and specific detection of tumor-associated miRNAs, aiding in early cancer diagnosis.
NAATs are invaluable tools for diagnosing genetic diseases, particularly for prenatal and pre-symptomatic diagnosis. Multiplex ddPCR allows for the non-invasive detection of fetal aneuploidies, while LAMP-based assays facilitate rapid sex determination in embryos. SNP typing methods, leveraging CRISPR-Cas systems, enable the identification of disease-associated genetic variations, paving the way for personalized medicine.
If you have related needs, please feel free to contact us for more information or product support.
Reference
| Cat.No | Product Name | Price |
|---|---|---|
| IEC-HMM-0013 | S-Adenosine Homocysteine Hydrolase, SAHH | Add To Cart |
| IEC-HMM-0045 | Uracil-DNA Glycosylase (UNG), Heat-labile | Add To Cart |
| IEC-HMM-0005 | Pyruvate Kinase, PK | Add To Cart |
| IEC-HMM-0034 | Proteinase K | Add To Cart |
| IEC-HMM-0033 | Cystathionine β-synthase, CBS | Add To Cart |
| IEC-HMM-0035 | Proteinase K Liquid (20 mg/mL) | Add To Cart |
| IEC-HMM-0012 | Homocysteine Methyltransferase, HMT | Add To Cart |
| IEC-HMM-0015 | S-adenosylmethionine Synthetase | Add To Cart |
| IEC-HMM-0027 | Mn-Superoxide Dismutase | Add To Cart |
| IEC-HMM-0041 | Uracil-DNA Glycosylase (UNG), E.coli | Add To Cart |
| IEC-HMM-0023 | Glutathione S-transferase | Add To Cart |
| IEC-HMM-0001 | AcyI-CoA Oxidase, ACO | Add To Cart |
| IEC-HMM-0031 | Malatedehydrogenase, MDH | Add To Cart |
| IEC-HMM-0014 | N-Acetylneuraminic Acid Aldolase, NAL | Add To Cart |
| IEC-HMM-0017 | Neuraminidase, NRH | Add To Cart |
This article is for research use only. Do not use in any diagnostic or therapeutic application.
|
There is no product in your cart. |