- Home
- Resource
- Explore & Learn
- Revolutionizing Clinical Diagnostics: RNA-Cleaving DNAzymes as Next-Gen Disease Detectors
- Home
- IVD
- By Technology Types
- By Diseases Types
- By Product Types
- Research
- Resource
- Distributors
- Company
RNA-cleaving DNAzymes (RCDs) represent a transformative class of functional nucleic acids that catalyze the site-specific cleavage of RNA substrates. Unlike antibodies or aptamers, RCDs combine high catalytic turnover, sequence-specific recognition, and chemical stability. Initially discovered in the mid-1990s for their metal ion-dependent cleavage activity, RCDs have rapidly evolved to target a wide range of clinically relevant analytes, including bacterial strains, protein biomarkers, small metabolites, and viral RNAs.
The 10–23 DNAzyme remains a cornerstone of this class. Dependent on Mg2+ ions, it efficiently cleaves all-RNA substrates at purine-pyrimidine junctions with catalytic constants approaching 10 min-1 under optimal conditions. This unparalleled catalytic rate positions RCDs as ideal components for point-of-care (POC) diagnostic assays.
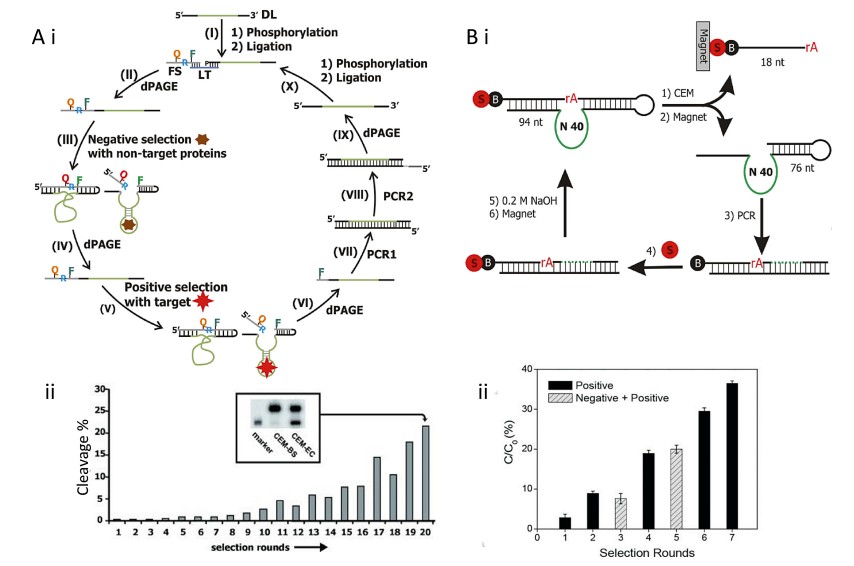 Fig.1 Overview of selection methods used to generate RNA-cleaving DNAzymes.(Ali M., et al., 2024)
Fig.1 Overview of selection methods used to generate RNA-cleaving DNAzymes.(Ali M., et al., 2024)

In Vitro Selection Methodologies
The core of RCD engineering lies in high-stringency in vitro selection, often using systematic evolution of ligands by exponential enrichment (SELEX). Starting with libraries comprising up to 1014 unique sequences, catalytically active species are enriched through positive selection (PS) and counter-selection (CS) cycles.
PAGE-based selection isolates cleaved products via polyacrylamide gel electrophoresis. In contrast, magnetic bead-based selection utilizes biotin-streptavidin interactions to immobilize the library, enabling efficient automation and reduced cycle times. These methods allow fine-tuned selection for target specificity and cleavage kinetics by adjusting pH, buffer conditions, and ion types.
Target Versatility and Specificity
RCDs have demonstrated activation by over 25 different metal ions, bacterial cell extracts, and individual proteins. The RFD-EC1 DNAzyme, for example, cleaves in the presence of E. coli lysate and discriminates against both gram-positive and gram-negative species. On the protein side, EPDz20 M5 is a landmark RCD selected directly against eosinophil peroxidase (EPX), a critical biomarker in asthma.
The hallmark of RCD-based detection is the transesterification reaction at RNA cleavage sites, generating distinct nucleic acid fragments. These products can be engineered with functional tags for signal transduction via:
To enhance sensitivity, RCDs are frequently coupled with isothermal amplification techniques. Rolling Circle Amplification (RCA), Loop-Mediated Isothermal Amplification (LAMP), and CRISPR-based methods have been successfully adapted to utilize RCD-cleaved fragments as primers or triggers.
A notable platform, REVEALR, integrates split DNAzymes with reverse transcription-recombinase polymerase amplification (RT-RPA) followed by T7 transcription to detect SARS-CoV-2 RNA at attomolar levels.
RCDs have shown promise in differentiating bacterial strains in complex matrices. For instance:
Table 1. Examples of Bacterial-Specific RCDs.
| RCD Identifier | Target Bacterium | Specificity | Detection Limit |
| RFD-EC1 | E. coli | Species level | 500 CFU/mL |
| RFD-CD1 | C. difficile (BI/027) | Strain-specific | Not evaluated |
| RFD-KP6 | Klebsiella pneumoniae | Species level, multiple strains | 105 CFU/mL |
Paper-based devices for H. pylori (DHp3T4) and S. aureus (RFD-SA6T1) incorporate urease- or gold nanoparticle-tagged DNA strands for visual readout. These devices demonstrate robust performance in matrices like stool and nasal mucus, requiring minimal processing and yielding results within 30 minutes.
The recent development of protein-activated DNAzymes marks a pivotal breakthrough. EPDz20 M5, derived via 15 PS rounds and CS steps against other eosinophil proteins, achieves nanomolar detection of EPX in unprocessed sputum.
MORAC (MOlecule Recognition based on Affinity and Catalysis) technology has further advanced the field by allowing simultaneous DNAzyme selection and target protein identification. For example, Dz04 was shown to bind apolipoprotein L6 (APOL6), and Dz41 recognized SARS1 in colon polyp tissues.
By fusing aptamer domains with catalytic cores, researchers have developed allosterically regulated RCDs for:
These constructs typically yield detection limits in the nanomolar to sub-nanomolar range and can be configured for fluorescence or colorimetric output.
The structure-specific recognition capability of RCDs also enables direct cleavage of highly structured RNA, such as microRNAs and viral genomes, providing a route for decentralized nucleic acid testing without reverse transcription.
RCD-based assays have been validated across diverse clinical matrices:
| Urine | RFD-EC1 and aRCD-EC1 achieved 100% sensitivity in UTI detection. |
| Sputum | EPDz20 M5 distinguished eosinophilic from non-eosinophilic asthma with 96–100% accuracy. |
| Tumor Tissue | AA12-5 differentiated malignant from benign breast tumors with 92% sensitivity. |
Challenges hindering commercialization include:
Strategies to address these issues involve buffer engineering (low pH, monovalent ions), use of nuclease-resistant analogs (e.g., FANA), and integration with automated processing systems.
To unlock the full potential of RCDs in diagnostics, several developments are underway:
RNA-cleaving DNAzymes have emerged as agile, precise, and powerful tools for next-generation clinical diagnostics. Their adaptability across platforms, compatibility with multiple biomarkers, and resilience in diverse biological matrices provide a unique foundation for the future of POC diagnostics. As technical challenges are addressed through innovation and integration, RCD-based assays are poised to become a cornerstone in molecular diagnostics, delivering rapid, reliable, and accessible solutions for global health.
If you have related needs, please feel free to contact us for more information or product support.
Reference
This article is for research use only. Do not use in any diagnostic or therapeutic application.

Cat.No. GP-DQL-00203
Rotavirus Antigen Group A and Adenovirus Antigen Rapid Test Kit (Colloidal Gold)

Cat.No. GP-DQL-00206
Adenovirus Antigen Rapid Test Kit (Colloidal Gold), Card Style

Cat.No. GP-DQL-00207
Adenovirus Antigen Rapid Test Kit (Colloidal Gold), Strip Style

Cat.No. GP-DQL-00211
Rotavirus Antigen Group A Rapid Test Kit (Colloidal Gold), Card Type

Cat.No. GP-DQL-00212
Rotavirus Antigen Group A Rapid Test Kit (Colloidal Gold), Card Type

Cat.No. IP-00189
Influenza A Rapid Assay Kit

Cat.No. GH-DQL-00200
Follicle-stimulating Hormone Rapid Test Kit (Colloidal Gold)
Cat.No. GH-DQL-00201
Insulin-like Growth Factor Binding Protein 1 Rapid Test Kit (Colloidal Gold)

Cat.No. GH-DQL-00202
Luteinizing Hormone Rapid Test Kit (Colloidal Gold)

Cat.No. GH-DQL-00208
Follicle-stimulating Hormone Rapid Test Kit (Colloidal Gold), Strip Style
Cat.No. GH-DQL-00209
Insulin-like Growth Factor Binding Protein 1 Rapid Test Kit(Colloidal Gold), Strip Style

Cat.No. GH-DQL-00210
Luteinizing Hormone Rapid Test Kit (Colloidal Gold), Strip Style
Cat.No. IH-HYW-0001
hCG Pregnancy Test Strip
Cat.No. IH-HYW-0002
hCG Pregnancy Test Cassette
Cat.No. IH-HYW-0003
hCG Pregnancy Test Midstream

Cat.No. GD-QCY-0001
Cocaine (COC) Rapid Test Kit

Cat.No. GD-QCY-0002
Marijuana (THC) Rapid Test Kit
Cat.No. GD-QCY-0003
Morphine (MOP300) Rapid Test Kit
Cat.No. GD-QCY-0004
Methamphetamine (MET) Rapid Test Kit
Cat.No. GD-QCY-0005
Methylenedioxymethamphetamine ecstasy (MDMA) Rapid Test Kit
Cat.No. GD-QCY-0006
Amphetamine (AMP) Rapid Test Kit
Cat.No. GD-QCY-0007
Barbiturates (BAR) Rapid Test Kit

Cat.No. GD-QCY-0008
Benzodiazepines (BZO) Rapid Test Kit
Cat.No. GD-QCY-0009
Methadone (MTD) Rapid Test Kit
Cat.No. GD-QCY-0011
Opiate (OPI) Rapid Test Kit
Cat.No. ID-HYW-0002
Multi-Drug Test L-Cup, (5-16 Para)
Cat.No. ID-HYW-0005
Multi-Drug Rapid Test (Dipcard & Cup) with Fentanyl
Cat.No. ID-HYW-0006
Multi-Drug Rapid Test (Dipcard & Cup) without Fentanyl
Cat.No. ID-HYW-0007
Multi-Drug 2~14 Drugs Rapid Test (Dipstick & Dipcard & Cup)
Cat.No. ID-HYW-0008
Fentanyl (FYL) Rapid Test (For Prescription Use)
Cat.No. ID-HYW-0009
Fentanyl Urine Test Cassette (CLIA Waived)
Cat.No. ID-HYW-0010
Fentanyl Urine Test Cassette (Home Use)
|
There is no product in your cart. |