- Home
- Resource
- Explore & Learn
- Allele-Specific PCR Using Fluorescent Probes: Guidelines for Primer Design in Genotyping Applications
- Home
- IVD
- By Technology Types
- By Diseases Types
- By Product Types
- Research
- Resource
- Distributors
- Company
Genetic variation lies at the heart of biological diversity, influencing everything from physical traits to disease susceptibility. Among the various forms of genetic variation, Single Nucleotide Polymorphisms (SNPs) stand out as the most abundant and versatile markers. SNPs are single base-pair differences scattered throughout the genome, occurring approximately once every 300 base pairs in humans. These minute variations can have profound effects on gene function, protein structure, and ultimately, phenotypic expression.
The detection and analysis of SNPs are crucial for understanding genetic predispositions to diseases, predicting drug responses, and advancing personalized medicine. Traditional methods such as whole-genome sequencing provide comprehensive genomic information but are often cost-prohibitive and time-consuming for targeted studies. This has led to the development of more efficient and cost-effective techniques, with Allele-Specific PCR (AS-PCR) using TaqMan probes emerging as a leading method for SNP genotyping.
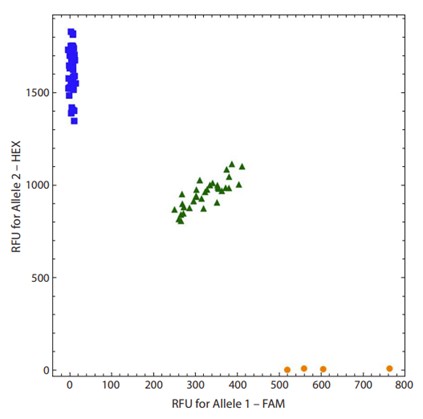 Fig.1 A high-quality allelic discrimination plot. (Devyatkin V. A., et al., 2024)
Fig.1 A high-quality allelic discrimination plot. (Devyatkin V. A., et al., 2024)
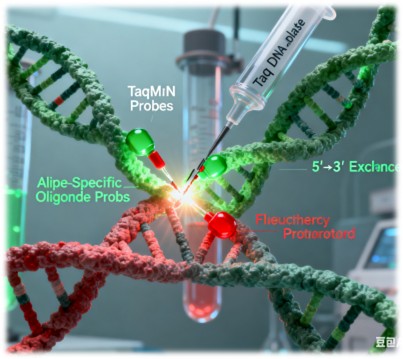
Allele-Specific PCR with TaqMan probes is a real-time PCR technique that enables the simultaneous amplification and detection of specific alleles. The method relies on the use of two allele-specific oligonucleotide probes, each labeled with a distinct fluorescent dye. These probes are designed to hybridize adjacent to the SNP site, with one probe complementary to the wild-type allele and the other to the mutant allele.
During PCR amplification, the Taq DNA polymerase extends the primers, and upon encountering the TaqMan probe, its 5'→3' exonuclease activity cleaves the probe, releasing the fluorescent reporter dye. The intensity of the fluorescence signal is proportional to the amount of PCR product generated, allowing for real-time monitoring of the reaction.
The key to allele discrimination lies in the specificity of probe hybridization. Probes are designed to have a higher melting temperature (Tm) when fully complementary to their target allele compared to when a mismatch exists. This difference in Tm ensures that only the fully complementary probe hybridizes efficiently, leading to the release of its fluorescent dye. In contrast, the probe with a mismatch remains largely intact, resulting in minimal fluorescence.
By comparing the fluorescence intensities of the two dyes, it is possible to determine the genotype of the sample. Homozygous samples will exhibit high fluorescence for only one dye, while heterozygous samples will show intermediate fluorescence levels for both dyes.


One of the primary advantages of AS-PCR with TaqMan probes is its high specificity and sensitivity. The method can accurately distinguish between alleles differing by a single nucleotide, even in complex genomic backgrounds. This is achieved through careful probe design, ensuring optimal hybridization conditions and minimizing non-specific binding.
Compared to whole-genome sequencing, AS-PCR with TaqMan probes is a rapid and cost-effective method for SNP genotyping. The technique requires minimal sample preparation and can be performed in a high-throughput manner, making it suitable for large-scale genetic studies. Additionally, the use of fluorescently labeled probes eliminates the need for post-PCR processing, further streamlining the workflow.
AS-PCR with TaqMan probes is highly versatile and can be adapted to various genetic markers and sample types. The method is compatible with DNA extracted from a wide range of sources, including blood, tissue, and saliva. Furthermore, the scalability of the technique allows for the simultaneous analysis of multiple SNPs, facilitating comprehensive genetic profiling.
If you have related needs, please feel free to contact us for more information or product support.
Reference
This article is for research use only. Do not use in any diagnostic or therapeutic application.
|
There is no product in your cart. |