- Home
- Resource
- Explore & Learn
- A Game-Changer in Respiratory Tract Infection Diagnosis
- Home
- IVD
- By Technology Types
- By Diseases Types
- By Product Types
- Research
- Resource
- Distributors
- Company
Respiratory tract infections (RTIs) remain a persistent global health challenge, contributing significantly to morbidity and mortality across all age groups. Lower respiratory tract infections, in particular, rank among the leading causes of death worldwide, with bacterial, viral, fungal, and atypical pathogens each playing distinct roles in disease pathogenesis. The clinical urgency of RTIs demands diagnostic tools that can rapidly identify causative agents, guide targeted therapy, and reduce the burden of inappropriate antibiotic use—a key driver of antimicrobial resistance.
Traditional diagnostic methods, reliant on microbial culture and biochemical identification, have long been the cornerstone of RTI pathogen detection. However, these approaches suffer from inherent limitations: slow turnaround times (often 3-5 days), low sensitivity for fastidious or non-culturable organisms, and inability to detect viruses or atypical pathogens without specialized assays. In contrast, metagenomic next-generation sequencing (mNGS) has emerged as a powerful alternative, offering broad-spectrum pathogen detection by sequencing all nucleic acids in a sample. Yet mNGS remains constrained by high costs, complex workflows, and lengthy analysis times, limiting its accessibility in routine clinical settings.
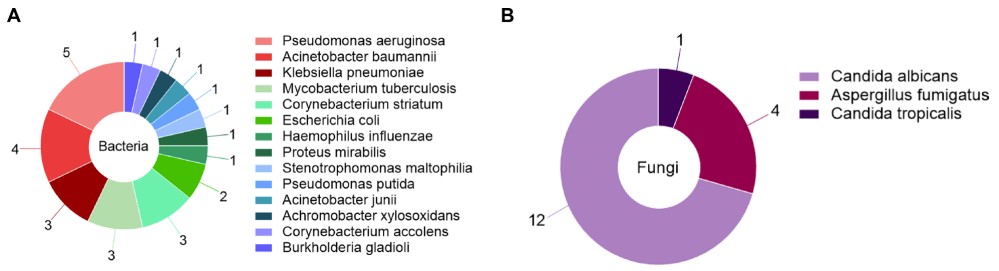 Fig.1 Bacteria and fungi were detected by traditional microbiological methods. A 28 bacteria were detected in group A. B 17 fungi were detected in group A. (Jiang J., et al., 2024)
Fig.1 Bacteria and fungi were detected by traditional microbiological methods. A 28 bacteria were detected in group A. B 17 fungi were detected in group A. (Jiang J., et al., 2024)
Against this backdrop, microfluidic-based in vitro diagnostic (IVD) technology has emerged as a transformative solution, combining the speed of molecular methods with the practicality of point-of-care testing. By miniaturizing and integrating sample processing, nucleic acid amplification, and detection into a single platform, microfluidic systems address the critical gaps in existing diagnostic pipelines.
A landmark study published in the Journal of Translational Medicine (2024) systematically evaluated three diagnostic approaches for RTI pathogen detection: traditional microbiological methods, mNGS, and microfluidic-based IVD technology. The study enrolled 300 patients with confirmed RTIs, randomized into three equal groups (n=100) based on the detection method used.
The study population had a mean age of 60.5 ± 16.1 years, with a near-equal gender distribution (160 males, 140 females). Clinical presentations were consistent across groups, with cough/expectoration (88–92%), wet rales (55–64%), and fever (29–37%) as the most common symptoms. Laboratory markers, including white blood cell count, neutrophil percentage, C-reactive protein, and procalcitonin, were comparable between groups, ensuring baseline equivalence.
All groups analyzed three clinical sample types: bronchoalveolar lavage fluid (BALF), pleural effusion, and blood. BALF and pleural effusion were stored at 4°C if delayed, while blood samples were processed immediately to avoid refrigeration artifacts. Each group followed distinct workflows:

Traditional methods (Group A): Specimens were plated on blood, chocolate, and Sabouraud agar, incubated for 24 hours, and colonies identified via Gram staining and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS).
mNGS (Group B): DNA extraction using commercial kits was followed by library construction, high-throughput sequencing, and bioinformatic alignment against reference genomes to identify pathogens.
Microfluidic-based IVD (Group C): A specialized chip kit capable of detecting 158 respiratory pathogens was used. Samples underwent rapid DNA extraction via centrifugation, loop-mediated isothermal amplification (LAMP) for 50 minutes, and automated data analysis, with a total workflow time of 1-4 hours.
Positive Detection Rates: Outperforming Traditional Methods
The primary endpoint—pathogen positive detection rate—revealed a stark contrast between methods. Traditional culture identified only 38% of positive samples, reflecting its well-documented limitations in detecting fastidious or non-culturable organisms. In comparison, mNGS achieved a 95% positive rate, while microfluidic-based IVD slightly outperformed mNGS at 96%. This difference held across all sample types:
| BALF | 39.5% (traditional) vs. 96.5% (mNGS) vs. 95.6% (microfluidic) |
| Pleural effusion | 25.0% (traditional) vs. 81.8% (mNGS) vs. 100% (microfluidic) |
| Blood | 33.3% (traditional) vs. 100% (mNGS) vs. 100% (microfluidic) |
Notably, microfluidic IVD detected a broader range of pathogens than traditional methods, which only identified bacteria and fungi. Like mNGS, microfluidic IVD detected viruses (e.g., Human gammaherpesvirus 4) and atypical pathogens (e.g., Mycoplasma pneumoniae), critical for diagnosing viral pneumonias and atypical infections that often mimic bacterial disease clinically.
Diagnostic Accuracy: Sensitivity and Specificity
Receiver Operating Characteristic (ROC) curve analysis quantified diagnostic performance, with the area under the curve (AUC) serving as a composite measure of sensitivity and specificity. Microfluidic IVD achieved the highest AUC (0.8968), followed by mNGS (0.8839) and traditional methods (0.6139). Key metrics included:
| Sensitivity | 95.12% (microfluidic) vs. 92.14% (mNGS) vs. 72.30% (traditional) |
| Specificity | 66.33% (microfluidic) vs. 62.50% (mNGS) vs. 55.35% (traditional) |
| Accuracy | 82% (microfluidic) vs. 80% (mNGS) vs. 61% (traditional) |
These results confirm that microfluidic IVD not only detects more pathogens but also minimizes false negatives, critical for ruling out infection in ambiguous cases.
Turnaround Time and Cost: Practical Advantages
In clinical settings, speed and cost directly impact patient outcomes and healthcare efficiency. Traditional culture required an average of 98.2 ± 24.3 hours, far too slow to guide acute management. mNGS reduced this to 13.5 ± 0.23 hours but remained costly at approximately 3500 Yuan per test. Microfluidic IVD, by contrast, delivered results in 2.45 ± 0.52 hours at half the cost of mNGS (1700 Yuan), making it feasible for high-volume use in busy hospitals.
Drug Resistance Gene Detection: Guiding Targeted Therapy
A unique advantage of microfluidic IVD is its ability to detect up to 35 critical drug resistance genes, including those encoding beta-lactamases and fluoroquinolone resistance. This capability addresses a major limitation of mNGS, which often requires additional assays to confirm resistance markers. By integrating resistance gene detection into the same workflow, microfluidic IVD provides actionable information for antibiotic stewardship, reducing trial-and-error prescribing and improving patient outcomes.

Bacterial Detection
Traditional methods detected 28 bacterial pathogens, with Pseudomonas aeruginosa (17.9%) and Acinetobacter baumannii (14.3%) as the most common. mNGS identified 154 bacterial species, including Mycobacterium tuberculosis (12.3%) and Haemophilus influenzae (5.2%). Microfluidic IVD detected 99 bacterial strains, with a focus on clinically relevant pathogens: Pseudomonas aeruginosa (11.1%), Klebsiella pneumoniae (9.1%), and Haemophilus influenzae (9.1%). While mNGS detected more total bacteria, microfluidic IVD prioritized species with high clinical impact, aligning with the needs of frontline clinicians.
Viral and Atypical Pathogens
Viruses and atypical pathogens are often missed by traditional methods but are critical drivers of RTIs. Microfluidic IVD detected 22 viral strains, including Human gammaherpesvirus 4 (31.8%) and Human alphaherpesvirus 1 (31.8%), and 17 atypical pathogens, dominated by Mycoplasma pneumoniae (78.6%). These results matched or exceeded mNGS performance, with the added benefit of faster identification—vital for antiviral or atypical pathogen-directed therapy (e.g., macrolides for Mycoplasma).
Microfluidic-based IVD technology represents a paradigm shift in RTI diagnostics, addressing the "triple challenge" of sensitivity, speed, and cost. Its integration into clinical workflows could:

While limitations exist—including potential nucleic acid contamination and the need for clinical correlation to interpret results—these are manageable with standardized protocols and training. Future iterations of microfluidic chips may expand pathogen panels, integrate host response markers, and further reduce costs, solidifying their role as a cornerstone of respiratory care.
In an era of emerging pathogens and antimicrobial resistance, microfluidic-based IVD technology is not merely an incremental improvement but a transformative tool—one that empowers clinicians to diagnose faster, treat smarter, and save more lives. Its adoption will undoubtedly redefine the standard of care for respiratory tract infections, making precision medicine a reality for patients worldwide.
If you have related needs, please feel free to contact us for more information or product support.
Reference
This article is for research use only. Do not use in any diagnostic or therapeutic application.

Cat.No. GP-DQL-00203
Rotavirus Antigen Group A and Adenovirus Antigen Rapid Test Kit (Colloidal Gold)

Cat.No. GP-DQL-00206
Adenovirus Antigen Rapid Test Kit (Colloidal Gold), Card Style

Cat.No. GP-DQL-00207
Adenovirus Antigen Rapid Test Kit (Colloidal Gold), Strip Style

Cat.No. GP-DQL-00211
Rotavirus Antigen Group A Rapid Test Kit (Colloidal Gold), Card Type

Cat.No. GP-DQL-00212
Rotavirus Antigen Group A Rapid Test Kit (Colloidal Gold), Card Type

Cat.No. IP-00189
Influenza A Rapid Assay Kit

Cat.No. GH-DQL-00200
Follicle-stimulating Hormone Rapid Test Kit (Colloidal Gold)
Cat.No. GH-DQL-00201
Insulin-like Growth Factor Binding Protein 1 Rapid Test Kit (Colloidal Gold)

Cat.No. GH-DQL-00202
Luteinizing Hormone Rapid Test Kit (Colloidal Gold)

Cat.No. GH-DQL-00208
Follicle-stimulating Hormone Rapid Test Kit (Colloidal Gold), Strip Style
Cat.No. GH-DQL-00209
Insulin-like Growth Factor Binding Protein 1 Rapid Test Kit(Colloidal Gold), Strip Style

Cat.No. GH-DQL-00210
Luteinizing Hormone Rapid Test Kit (Colloidal Gold), Strip Style
Cat.No. IH-HYW-0001
hCG Pregnancy Test Strip
Cat.No. IH-HYW-0002
hCG Pregnancy Test Cassette
Cat.No. IH-HYW-0003
hCG Pregnancy Test Midstream

Cat.No. GD-QCY-0001
Cocaine (COC) Rapid Test Kit

Cat.No. GD-QCY-0002
Marijuana (THC) Rapid Test Kit
Cat.No. GD-QCY-0003
Morphine (MOP300) Rapid Test Kit
Cat.No. GD-QCY-0004
Methamphetamine (MET) Rapid Test Kit
Cat.No. GD-QCY-0005
Methylenedioxymethamphetamine ecstasy (MDMA) Rapid Test Kit
Cat.No. GD-QCY-0006
Amphetamine (AMP) Rapid Test Kit
Cat.No. GD-QCY-0007
Barbiturates (BAR) Rapid Test Kit

Cat.No. GD-QCY-0008
Benzodiazepines (BZO) Rapid Test Kit
Cat.No. GD-QCY-0009
Methadone (MTD) Rapid Test Kit
Cat.No. GD-QCY-0011
Opiate (OPI) Rapid Test Kit
Cat.No. ID-HYW-0002
Multi-Drug Test L-Cup, (5-16 Para)
Cat.No. ID-HYW-0005
Multi-Drug Rapid Test (Dipcard & Cup) with Fentanyl
Cat.No. ID-HYW-0006
Multi-Drug Rapid Test (Dipcard & Cup) without Fentanyl
Cat.No. ID-HYW-0007
Multi-Drug 2~14 Drugs Rapid Test (Dipstick & Dipcard & Cup)
Cat.No. ID-HYW-0008
Fentanyl (FYL) Rapid Test (For Prescription Use)
Cat.No. ID-HYW-0009
Fentanyl Urine Test Cassette (CLIA Waived)
Cat.No. ID-HYW-0010
Fentanyl Urine Test Cassette (Home Use)
|
There is no product in your cart. |